Nuclear Transport in a Divergent Eukaryotes
Eukaryogenesis is a major evolutionary transition in the history of life, that led to the emergence of extensive intracellular compartmentalization within cells, and thus eukaryotes. Much of the core architecture of the eukaryotic cell was established well over one billion years ago. Significantly, many cellular systems possess lineage-specific features, and architectural and compositional variation of complexes and pathways are likely keyed to specific functional adaptations. To reconstruct eukaryogenesis and the pathways that lead to and from the prokaryote/eukaryote transition, we must look for the imprints that this process left at the structural and molecular levels in the architecture of protein assemblies in modern-day conserved eukaryotic molecular machines, such as the NPC, a ubiquitous eukaryotic mega-machine responsible for mediating and regulating flux between the nucleus and the cytoplasm. However, most of our knowledge of the NPC structure are function are derived from decades of work in yeast and vertebrates, both members of the Opisthokonta, one of five or six major supergroups of the eukaryotic lineage.
Thus, to expand our understand and trace some of the evolutionary history of the NPC, we turned to trypanosomes, a diverse family of parasitic protozoans that are obligatory parasites of invertebrates, vertebrates, and plants, and thus cause major public health and economic problems in the developing world. cause major public health and economic problems in the developing world and more recently in the United States (Chagas disease). Trypanosomes are members of the Excavata supergroup, early diverging eukaryotes that exhibit biological traits that were once considered unique to trypanosomes, but that have now proven to be shared amongst other eukaryotic lineages. Indeed, several key features of molecular biology were first identified in trypanosomes. Examples include antigenic variation, GPI-anchored proteins, RNA editing, polycistronic transcription and trans-splicing.
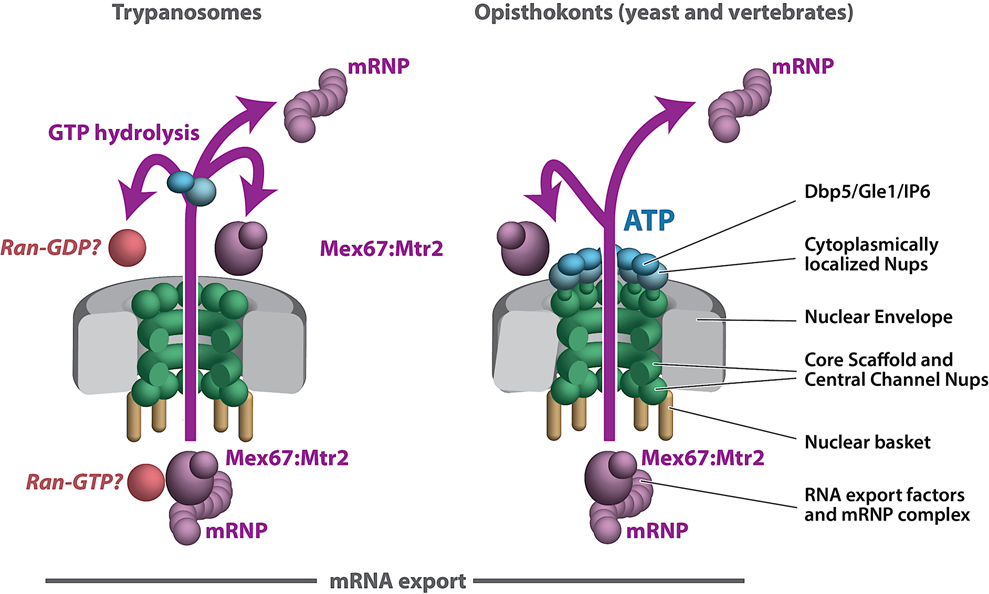
We are exploiting trypanosome parasites to investigate NPC evolution and conservation at the level of protein–protein interactions and composition. We have found that the NPC structural scaffold is generally conserved, albeit with lineage-specific elements. However, there are major differences in the organization and architecture of key NPC modules in the highly divergent eukaryote Trypanosoma brucei, including significant variation in pore membrane proteins and an alternate mode of mRNA export that differs significantly with the text book mechanisms described in fungi and vertebrates (Opisthokonts). The trypanosome NPC architecture highlights the fact that we do not fully know if the textbook description of RNA export is universal to all eukaryotes or genus specific, nor if all levels of RNA export control in Opisthokonts are fully understood. This underscores Theodosius Dobzhansky’s concept that “nothing in biology makes sense except in the light of evolution”, with discoveries in trypanosome cell biology repeatedly shedding extremely valuable light on the elasticity of cell biology principles and mechanisms conserved across all eukaryotes.
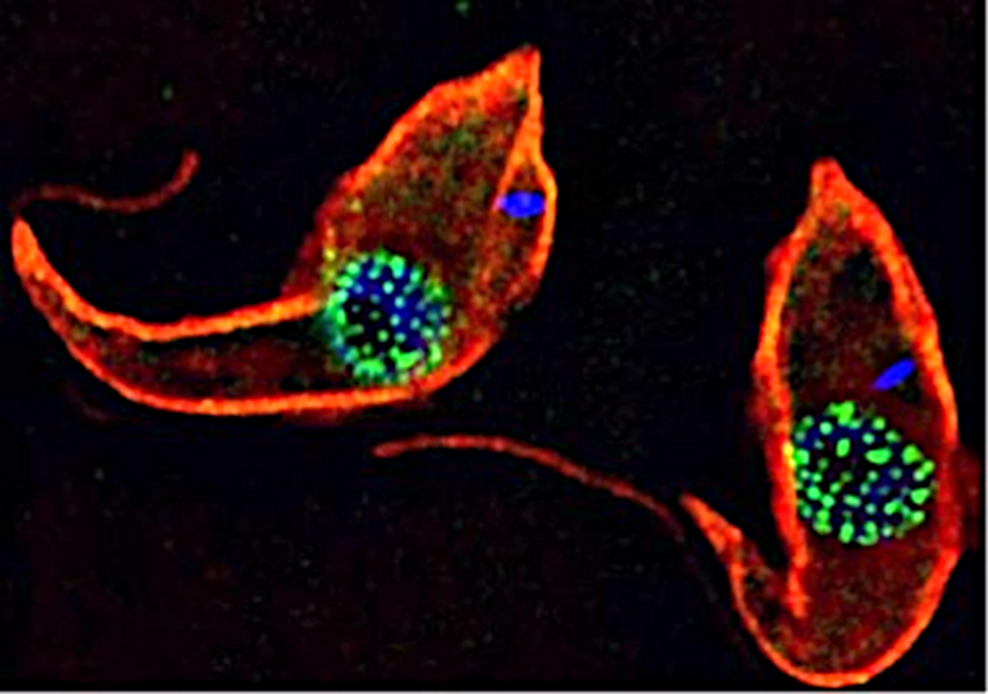
Trypanosomes as models for the study of nuclear pore complex (NPC) structure function and evolution. Trypanosomes are unicellular flagellated protozoan parasites. Trypanosomes are encased in a subpellicular tubulin array (red – anti tubulin antibody (KMX1)). TbNup89, a nucleoporin component of the NPC has been in situ tagged with GFP to form a TbNup89-GFP chimeric protein (green). DNA is stained blue (DAPI). The DNA outside the nucleus (blue) is the kinetoplast which is the mitochondrial DNA in these organisms.